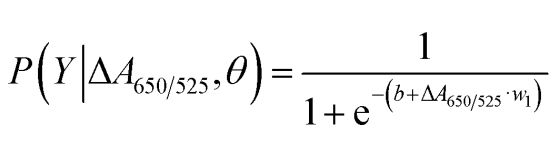Early detection of cancer based on liquid biopsy is a new direction of cancer detection and diagnosis proposed by the US National Cancer Institute in recent years, with the aim of detecting early cancer or even precancerous lesions. It has been widely used as a novel biomarker for the early diagnosis of various malignancies, including lung cancer, gastrointestinal tumours, gliomas and gynaecological tumours.
The emergence of platforms to identify methylation landscape (Methylscape) biomarkers has the potential to significantly improve existing early screening for cancer, putting patients at the earliest treatable stage.
Recently, researchers have developed a simple and direct sensing platform for methylation landscape detection based on cysteamine decorated gold nanoparticles (Cyst/AuNPs) combined with a smartphone-based biosensor that enables rapid early screening of a wide range of tumours. Early screening for leukaemia can be performed within 15 minutes after DNA extraction from a blood sample, with an accuracy of 90.0%. Article title is Rapid detection of cancer DNA in human blood using cysteamine-capped AuNPs and a machine learning-enabled smartphone。
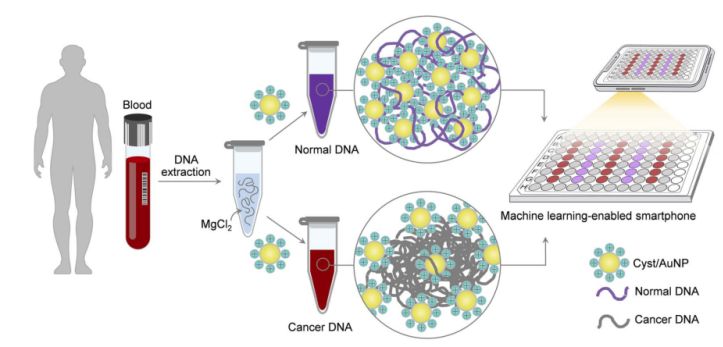
Figure 1. A simple and fast sensing platform for cancer screening via Cyst/AuNPs components can be accomplished in two simple steps.
This is shown in Figure 1. First, an aqueous solution was used to dissolve the DNA fragments. Cyst/AuNPs were then added to the mixed solution. Normal and malignant DNA have different methylation properties, resulting in DNA fragments with different self-assembly patterns. Normal DNA aggregates loosely and eventually aggregates Cyst/AuNPs, which results in the red-shifted nature of Cyst/AuNPs, so that a change in colour from red to purple can be observed with the naked eye. In contrast, the unique methylation profile of cancer DNA leads to the production of larger clusters of DNA fragments.
Images of 96-well plates were taken using a smartphone camera. Cancer DNA was measured by a smartphone equipped with machine learning compared to spectroscopy-based methods.
Cancer screening in real blood samples
To extend the utility of the sensing platform, the investigators applied a sensor that successfully distinguished between normal and cancerous DNA in real blood samples. methylation patterns at CpG sites epigenetically regulate gene expression. In almost all cancer types, changes in DNA methylation and thus in the expression of genes that promote tumourigenesis have been observed to alternate.
As a model for other cancers associated with DNA methylation, the researchers used blood samples from leukaemia patients and healthy controls to investigate the effectiveness of the methylation landscape in differentiating leukaemic cancers. This methylation landscape biomarker not only outperforms existing rapid leukaemia screening methods, but also demonstrates the feasibility of extending to early detection of a wide range of cancers using this simple and straightforward assay.
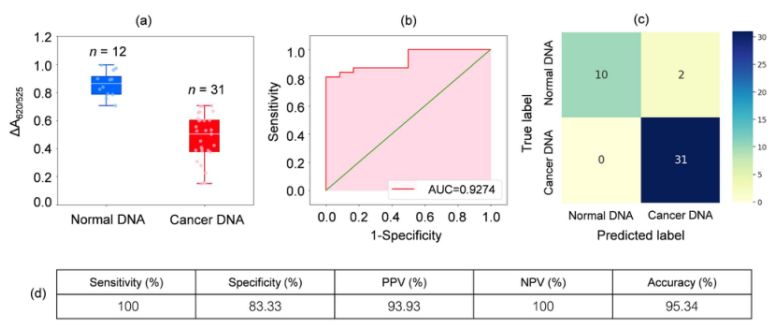
DNA from blood samples from 31 leukaemia patients and 12 healthy individuals was analysed. as shown in the box plot in Figure 2a, the relative absorbance of the cancer samples (ΔA650/525) was lower than that of DNA from normal samples. this was mainly due to the enhanced hydrophobicity leading to dense aggregation of cancer DNA, which prevented the aggregation of Cyst/AuNPs. As a result, these nanoparticles were completely dispersed in the outer layers of the cancer aggregates, which resulted in a different dispersion of Cyst/AuNPs adsorbed on normal and cancer DNA aggregates. ROC curves were then generated by varying the threshold from a minimum value of ΔA650/525 to a maximum value.
Figure 2.(a) Relative absorbance values of cyst/AuNPs solutions showing the presence of normal (blue) and cancer (red) dna under optimized conditions
(DA650/525) of box plots; (b) ROC analysis and evaluation of diagnostic tests. (c) Confusion matrix for the diagnosis of normal and cancer patients. (d) Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and accuracy of the developed method.
As shown in Figure 2b, the area under the ROC curve (AUC = 0.9274) obtained for the developed sensor showed high sensitivity and specificity. As can be seen from the box plot, the lowest point representing the normal DNA group is not well separated from the highest point representing the cancer DNA group; therefore, logistic regression was used to differentiate between the normal and cancer groups. Given a set of independent variables, it estimates the probability of an event occurring, such as a cancer or normal group. The dependent variable ranges between 0 and 1. The result is therefore a probability. We determined the probability of cancer identification (P) based on ΔA650/525 as follows.
where b=5.3533,w1=-6.965. For sample classification, a probability of less than 0.5 indicates a normal sample, while a probability of 0.5 or higher indicates a cancer sample. Figure 2c depicts the confusion matrix generated from the leave-it-alone cross-validation, which was used to validate the stability of the classification method. Figure 2d summarises the diagnostic test evaluation of the method, including sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV).
Smartphone-based biosensors
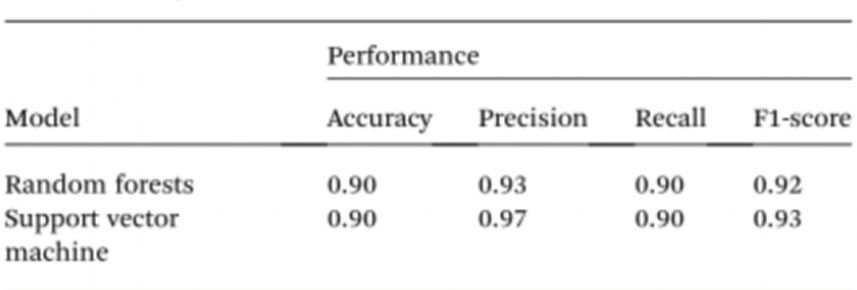
To further simplify sample testing without the use of spectrophotometers, the researchers used artificial intelligence (AI) to interpret the colour of the solution and distinguish between normal and cancerous individuals. Given this, computer vision was used to translate the colour of the Cyst/AuNPs solution into normal DNA (purple) or cancerous DNA (red) using images of 96-well plates taken through a mobile phone camera. Artificial intelligence can reduce costs and improve accessibility in interpreting the colour of nanoparticle solutions, and without the use of any optical hardware smartphone accessories. Finally, two machine learning models, including Random Forest (RF) and Support Vector Machine (SVM) were trained to construct the models. both the RF and SVM models correctly classified the samples as positive and negative with an accuracy of 90.0%. This suggests that the use of artificial intelligence in mobile phone-based biosensing is quite possible.
Figure 3.(a) Target class of the solution recorded during the preparation of the sample for the image acquisition step. (b) Example image taken during the image acquisition step. (c) Colour intensity of the cyst/AuNPs solution in each well of the 96-well plate extracted from the image (b).
Using Cyst/AuNPs, researchers have successfully developed a simple sensing platform for methylation landscape detection and a sensor capable of distinguishing normal DNA from cancer DNA when using real blood samples for leukaemia screening. The developed sensor demonstrated that DNA extracted from real blood samples was able to rapidly and cost-effectively detect small amounts of cancer DNA (3nM) in leukaemia patients in 15 minutes, and showed an accuracy of 95.3%. To further simplify sample testing by eliminating the need for a spectrophotometer, machine learning was used to interpret the colour of the solution and differentiate between normal and cancerous individuals using a mobile phone photograph, and accuracy was also able to be achieved at 90.0%.
Reference: DOI: 10.1039/d2ra05725e
Post time: Feb-18-2023
 中文网站
中文网站